Blog
Examining Brain Complexity: How Single-Cell and Spatial Transcriptomics Advance Neuroscience Discoveries
By Courtney Nirenberchik, Marketing Manager, Signios Bio
The Ongoing Quest in Brain Health
June is Alzheimer’s & Brain Awareness Month, a key time to consider the significant challenges posed by neurological disorders. From Alzheimer’s and Frontotemporal Dementia (FTD) to Traumatic Brain Injury (TBI) and rare conditions like Friedreich’s Ataxia, the brain’s complexity requires innovative research approaches. At Signios Bio, we are focused on aiding scientists by providing state-of-the-art omics technologies and expert guidance. What we’ve found is single-cell transcriptomics and spatial transcriptomics, specifically, are offering detailed insights into the cellular and molecular foundations of brain health and disease, which can accelerate the path towards new diagnostics and therapeutics.
Clarifying Cellular Landscapes With Single-Cell & Spatial Insights
Understanding how individual cells and their microenvironments respond to disease or injury is fundamental. Signios Bio has supported important research that illustrates the utility of these high-resolution genomic techniques.
Detailing Neuroinflammation and Repair in Traumatic Brain Injury (TBI): In collaboration with Professor Michelle H. Theus of Virginia Tech, Signios provided crucial single-cell and spatial transcriptomics support for TBI research in murine models. Spatial transcriptomics allowed for detailed mapping of injured versus uninjured brain regions, visually demonstrating how cell populations like astrocytes and endothelial cells adapt within the lesion site. This approach offered a closer look at processes such as blood-brain barrier breakdown and inflammation. Complementing this, single-cell RNA sequencing (scRNA-seq) identified specific gene expression changes in astrocytes, important for understanding neuroinflammation and regeneration, while also providing clues about vascular repair mechanisms within endothelial cells. Comparing different mouse models, these technologies pinpointed physiological and cellular changes, including shifts in astrocyte and endothelial cell populations within the injured area.
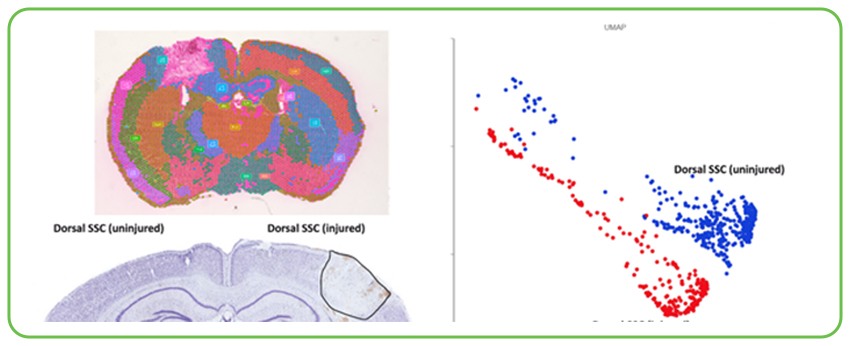
Spatial transcriptomics data from a murine Traumatic Brain Injury (TBI) model contrasts injured and uninjured brain regions, visualizing how specific cell populations adapt and gene expression patterns shift within and around the injury site. This level of spatial resolution is key to understanding the localized cellular responses, including blood-brain barrier dynamics and neuroinflammation, following trauma.
Characterizing Therapeutic Cell Replacement in Progranulin Deficiency (FTD/CLN11): Progranulin (GRN) deficiency leads to serious neurodegenerative conditions like FTD and Neuronal Ceroid Lipofuscinosis type 11 (CLN11). Research from Stanford University, with scRNA-seq and bioinformatic services from Signios, investigated an innovative conditioning regimen to enable robust CNS repopulation by hematopoietic-derived microglia-like cells (MGLCs). Single-cell RNA sequencing was instrumental in characterizing the engrafted MGLCs, showing a heterogeneity with six distinct subpopulations. This detailed analysis showed that MGLCs acquire a hybrid transcriptional identity, distinct from both embryonic microglia and bone marrow cells, and that the conditioning regimen induces specific local cytokine signals (like CSF1, IL34, and various CCL chemokines) important for successful engraftment. This approach restored GRN levels and normalized lipid metabolism in a Grn-/- mouse model, demonstrating therapeutic potential.
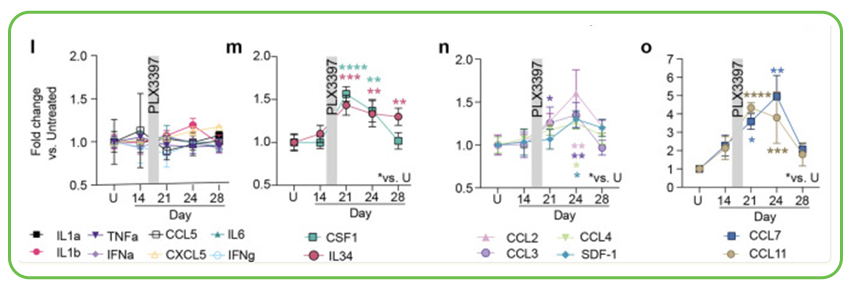
Serial brain cytokine analysis showing transient changes in specific cytokines and chemokines (such as CSF1, IL34, CCL2, CCL7, CCL11) in the brain following an optimized conditioning regimen involving Busulfan and PLX3397. These localized signaling events are thought to contribute to the effective recruitment and repopulation of the microglial niche by therapeutic bone marrow-derived cells.
Studying Disease Mechanisms and Therapeutic Pathways
Beyond cellular responses to injury and replacement, single-cell and spatial technologies are key to dissecting fundamental disease mechanisms.
Investigating Mitochondrial Dynamics in Friedreich’s Ataxia (FA): Friedreich’s Ataxia is a serious mitochondrial disease with no cure. A study from Stanford University, supported by Signios for its scRNA-seq components, utilized myeloid cell replacement therapy in an FA mouse model. This research showed that the therapy facilitated the transfer of healthy mitochondria from donor myeloid cells to various CNS cells. Subsequent scRNA-seq analysis of sorted brain cells (based on mitochondrial uptake) pinpointed significant transcriptomic changes. Cells that received mitochondria showed upregulation of genes involved in ATP synthesis and oxidative phosphorylation, indicating metabolic improvement. The benefits extended to cardiac function, where macrophage replacement improved mitochondrial activity and partially reversed cardiomyopathy, as also supported by bulk RNA sequencing of heart tissue.

Fluorescence microscopy images showing FA fibroblasts (larger cells) that have taken up mKate2-labeled mitochondria (red signal) after being cultured with donor macrophages (smaller, green GFP-expressing cells, with red mitochondria). This visual evidence supports the hypothesis that intercellular mitochondrial transfer is a mechanism by which myeloid cells can deliver healthy mitochondria to FA-affected cells, potentially restoring metabolic function.
Exploring APOE’s Role in Alzheimer’s Disease Microglial Response: The APOE gene is a major genetic risk factor for Alzheimer’s disease (AD). Research published in Nature Communications, for which Signios provided scRNA-seq sequencing services, investigated how different APOE isoforms (E3 vs. E4) influence microglial interaction with amyloid-β (Aβ). The study found that APOE3 lipoproteins, unlike APOE4, induced faster microglial migration towards Aβ and facilitated better Aβ uptake in preclinical AD models. Single-cell RNA sequencing was important in demonstrating that APOE3 lipoprotein infusion upregulated a higher proportion of genes linked to an activated microglia response compared to APOE4. This isoform-specific effect on microglial transcriptomes and function offers important information for developing targeted AD therapies.
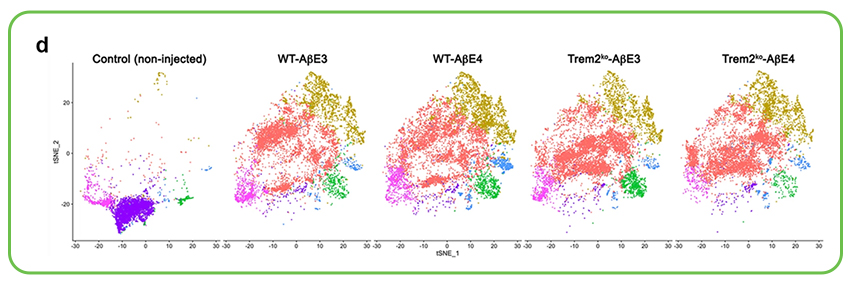
A series of t-SNE plots from single-cell RNA sequencing illustrating the distribution of distinct microglial cell clusters in response to cortical infusions of Aβ complexed with either APOE3 or APOE4 lipoproteins in wild-type (WT) and TREM2 knockout (Trem2ko) preclinical Alzheimer’s disease models, compared to non-injected controls. These visualizations highlight how APOE isoform and TREM2 status significantly alter the microglial landscape and activation states upon Aβ challenge.
Signios Biosciences: Your Partner in Neurological Discovery
The complexity of the brain requires multifaceted approaches. Signios Bio provides a suite of solutions for neuroscience research:
- Our Neuroregulation Signal service identifies changes in methylation and chromatin structure, and vulnerable neuronal populations using transcriptomics and epigenomics at single-cell and spatial resolution.
- The Neuroimmune Signal service characterizes disease-associated neural cell populations and maps immune cell infiltration in the brain via spatial transcriptomics and epigenomics.
- For challenges like repeat expansion disorders (e.g., Friedreich’s Ataxia, Huntington’s), our highly accurate long-read sequencing services provide the necessary resolution to characterize these complex genomic regions.
As Professor Theus remarked about her TBI research, the journey from early transcriptomic insights to testing drug therapies is achievable. Signios is dedicated to providing the genomic tools and expertise to aid such developments.
Let’s examine the complexities of the brain, together. Contact us today to learn how our tailored omics solutions can assist your neuroscience research.
