RNA Sequencing Services
Decode molecular mechanisms through high-quality gene expression profiling
Cost efficient gene expression data from both good and poor quality RNA samples
RNA sequencing services designed to support a broad range of transcriptomic studies across sample types and research areas.
Signios Bio provides high-quality RNA sequencing (RNA-seq) services optimized for diverse research applications. Our flexible workflows support mRNA and total RNA captures, low-input material and FFPE sample processing, helping you generate reproducible results for every stage of research—from exploratory studies to advanced therapeutic programs and beyond.

Why Choose Signios Bio for RNA Sequencing?
![]()
End-to-End Support
From experimental design to bioinformatics analysis and reporting.
![]()
Multi-Omics Integration
Seamlessly combine RNA-seq data with epigenetic, immune profiling, single-cell, spatial, or proteomics datasets.
![]()
Fast Turnaround Times
Optimized protocols for efficient sample processing and data delivery.
![]()
Flexible Sample Types
Accepted inputs include total RNA, tissue, cells, FFPE samples, and biofluids.
![]()
Trusted by Leading Research Institutions
Supporting oncology, immunology, neuroscience, and drug development projects.
RNA Sequencing Workflows
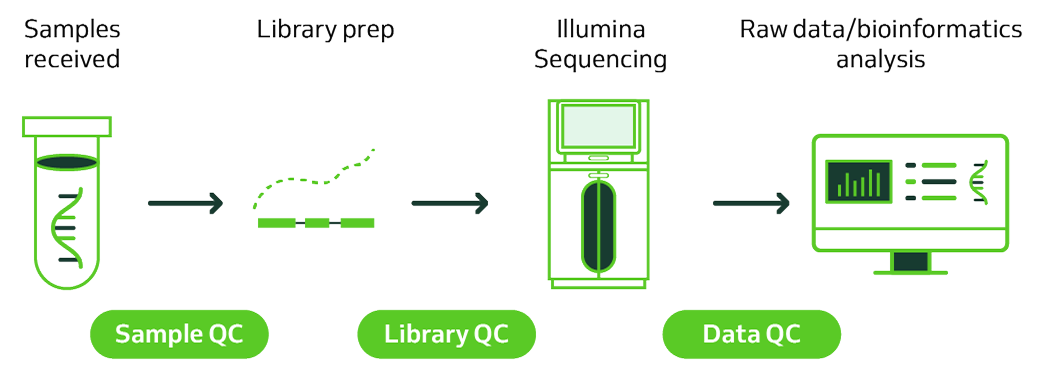
Library Prep
Suitable for
Starting Material
Input Amount
Library Prep
Illumina Stranded mRNA
Suitable for
High-quality RNA from any source, to capture stranded mRNA information
Starting Material
Tissues, cells, blood
and total RNA from mammalian and species with polyA
Input Amount
50 ng 1 μg RNA
Library Prep
KAPA mRNA HyperPrep
Suitable for
High-quality RNA from any source, to capture stranded mRNA information
Starting Material
Tissues, cells, blood and Total RNA from mammalian and
other species with polyA
Input Amount
100 ng – 1 μg
RNA
Library Prep
Takara SMART-Seq mRNA
Suitable for
Capture mRNA information from low total RNA yields and low
Starting Material
Total RNA, cells
Input Amount
10 pg – 10 ng RNA, 10,000 cells
Library Prep
Takara Pico v3
Suitable for
Capture stranded total RNA information from ultra-low input and FFPE sources
Starting Material
Total RNA
Input Amount
250 pg – 10 ng RNA
Library Prep
KAPA RNA HyperPrep with RiboErase Globin
Suitable for
Blood
Starting Material
Blood
Input Amount
3 mL
Accepted Sample Types for RNA Sequencing
Sample Type
Requirements
Storage/Shipping
Notes
Sample Type
Total RNA
Requirements
≥ 10-1000 ng
RIN ≥ 0-7 recommended
Storage/Shipping
Dry ice
Ideal Applications
Specialized handling of RNA from all sources
Sample Type
Tissue
Requirements
20–50 mg
Storage/Shipping
Flash-frozen or RNA later
Ideal Applications
Fresh or frozen tissue
Sample Type
Cells
Requirements
1–1 million cells
Storage/Shipping
Dry ice
Ideal Applications
Fresh, Frozen or fixed cells. Collection protocols can be provided
Sample Type
FFPE Samples
Requirements
≥ 5 sections (10 μm each)
DV200≥ 20%
Storage/Shipping
Room temperature
Ideal Applications
Blocks, curls and slides accepted
Sample Type
Biofluids (plasma, serum)
Requirements
200–500 μL
Storage/Shipping
Dry ice
Ideal Applications
For exosomal/small RNA studies
Sample Type
Blood
Requirements
3 mL
Storage/Shipping
On ice 4c
Ideal Applications
Collect in EDTA tube
Not sure if your sample type qualifies? Our team can advise on collection methods and submission requirements.
Comprehensive Bioinformatics for RNA-Seq
Every project includes expert bioinformatics analysis with customizable reporting:
- Quality control & read alignment
- Transcript quantification & differential expression analysis
- Pathway enrichment & gene set analysis
- Interactive reports featuring heatmaps, volcano plots, and PCA plots
- Integration with single-cell and proteomics data (upon request)
Applications of RNA Sequencing
![]()
Gene Expression Profiling
![]()
Biomarker Discovery
![]()
Drug Mechanism of Action Studies
![]()
Non-Coding RNA Research
![]()
Disease Pathway Analysis
![]()
Multi-Omics Systems Biology
Get Started with RNA Sequencing
Our team is ready to support your project from start to finish. Whether you’re working with challenging samples or large study cohorts, Signios Bio delivers reliable RNA-seq data with expert guidance.
Frequently Asked Questions
1. How do I decide between poly(A) selection and rRNA depletion for my RNA-Seq project?
Use poly(A) selection for high-quality, eukaryotic RNA focused on coding transcripts. Choose rRNA depletion when working with degraded RNA (e.g., FFPE), total RNA, or if your goal includes studying non-coding RNAs, pre-mRNAs, or bacterial/viral RNA.
2. What’s the minimum RNA input required, and can I sequence low-input or rare samples?
Signios Bio supports RNA-Seq with as little as 1 ng of total RNA using optimized low-input protocols. Single-cell or rare cell populations can also be sequenced using SMART-Seq or 10x Genomics platforms.
3. How important is RNA integrity, and can you work with FFPE samples?
Yes — but RNA integrity is critical. FFPE samples often have degraded RNA (low RIN scores), requiring special extraction and library preparation protocols. Signios Bio team can perform QC and suggest rRNA depletion-based or 3’-end counting methods for success.
4. How do stranded RNA-Seq libraries benefit transcriptome analysis?
Stranded libraries preserve transcript directionality, which improves gene quantification accuracy, especially for antisense RNAs, overlapping genes, and fusion transcripts. It’s especially valuable for cancer transcriptomics and viral transcript mapping.
5. What read length and depth do I need for meaningful results?
- Gene expression: 20–30 M reads/sample (paired-end 2 x 75 or 2 x 100)
- Alternative splicing: 50 M+ reads/sample
- Transcript discovery/novel isoforms: 100 M+
- Signios Bio offers flexible platforms based on Illumina and long-read sequencing (as needed).
6. Can RNA-Seq detect gene fusions, novel isoforms, and transcript variants?
Yes. Paired-end reads and strand-specific protocols help identify gene fusions, chimeric transcripts, and splicing events. Long-read sequencing options (ONT or PacBio) are ideal for full-length isoform detection.
7. Do I need to remove globin mRNA or mitochondrial RNA for blood/tissue samples?
Yes — globin mRNA in blood and mitochondrial RNA in certain tissues can dominate libraries. Custom depletion protocols are recommended and available through Signios Bio to improve transcriptome coverage.
8. Can you help with downstream analysis if I don’t have a bioinformatics team?
Absolutely. Signios Bio offers end-to-end RNA-Seq solutions, including QC, alignment, expression quantification, differential expression analysis, and custom reporting — tailored to your experimental design and organism.
9. How is your RNA-Seq pipeline different from larger commercial providers?
Signios Bio combines scientific consultation, project customization, and direct communication with the lab team. Unlike templated pipelines, we offer flexible protocols, QC transparency, and client-specific optimization for academic and biotech clients.
10. How long does RNA-Seq take, from sample submission to data delivery?
Turnaround time varies by project scale, but typical RNA-Seq projects are completed in 2–4 weeks, including QC, library prep, sequencing, and bioinformatics. Faster turnaround options are available on request.
11. Can I integrate RNA-Seq with other omics services you offer?
Yes. Signios supports multi-omics integration, including RNA-Seq + DNA-Seq, methylation, or proteomics to support systems biology, biomarker discovery, and drug development workflows.
Contact Us

