Sequencing Services
Uncover Genetic Signatures
Finding the genetic drivers of disease requires you to ask new kinds of questions and use the latest sequencing technologies. Vast amounts of complex experimental data must then be translated into knowledge that informs the next hypothesis and advances your journey toward breakthrough discoveries.
Explore our sequencing solutions to find exactly what you need.
Access the Latest Technologies
Illumina
Novaseq X Plus
Illumina
MiSeq

PacBio
Revio
10X Genomics
Chromium
10X Genomics
Visium
Sony
MA900 Cell Sorter
Premade Library Sequencing
Unparalleled data quality, versatile read lengths and comprehensive support
Send us your Illumina sequencing libraries for various applications including single cell, transcriptomics, WGS, WES, amplicon, bi-sulfite, TCR, BCR and more. Our trusted genomic solutions are tailored to meet your specific needs. You can receive FASTQ files or opt for our cutting-edge bioinformatics solutions.
Unparalleled Data Quality
We’ve completed thousands of sequencing projects and have had results published in high impact peer-reviewed journals.
24/7 Access to our Online Portal
Enjoy the convenience of uploading manifests and registering libraries through our user-friendly customer portal, NxgenLIMS.
Versatile Read Lengths
Tailor your sequencing needs with options like PE250, PE150, PE100, PE50, and customizable read lengths.
Cost-Effective with Rapid Turnaround
Experience the dual benefits of affordability and rapid project completion.
Express Sequencing Service
Need results in a hurry? Use our expedited service for an even faster turnaround.
Flexible Lane/Flow Cell Choices
Customize your sequencing project with flexible choices for lanes and flow cells.
Comprehensive Support
Engage with us scientist-to-scientist for quality control (QC) and bioinformatics support.
Request premade sequencing services
RNA Sequencing
Reveal molecular mechanisms through high-quality gene expression profiling
We deliver valuable gene expression data from both good and poor-quality RNA samples. Flexible library preparation options are based on RNA quality and quantity, allowing researchers to generate optimal-quality data from their samples. We can work with RNA from all sources such as FFPE or low cell counts by leveraging highly optimized workflows and a consultative approach with customers to make informed decisions.
RNA Sequencing Services
What library prep workflows are available for RNA sequencing?
Library
Prep
Suitable
for
Starting
Material
Input
Amount
Library Prep
Illumina Stranded mRNA
Suitable for
High-quality RNA from any source, to capture stranded mRNA information
Starting Material
Tissues, cells, blood and total RNA from mammalian and species with polyA
Input Amount
50ng – 1ug RNA
Library Prep
KAPA mRNA HyperPrep
Suitable for
High-quality RNA from any source, to capture stranded mRNA information
Starting Material
Tissues, cells, blood and Total RNA from mammalian and other species with polyA
Input Amount
100ng – 1ug RNA
Library Prep
Takara SMART-Seq mRNA
Suitable for
Capture mRNA information from low total RNA yields and low starting cell counts
Starting Material
Total RNA, cells
Input Amount
10pg – 10ng RNA, 10,000 cells
Library Prep
Takara Pico v3
Suitable for
Capture stranded total RNA information from ultra-low input and FFPE sources
Starting Material
Total RNA
Input Amount
250pg – 10ng RNA
Library Prep
KAPA RNA HyperPrep with RiboErase Globin
Suitable for
Blood
Starting Material
Blood
Input Amount
3mL
What bioinformatics services are available for RNA sequencing?
Basic Analysis
- Data QC report
- Raw data FASTQ file
Standard Analysis
- All items in Basic Analysis +
- Alignment (BAM) file
- Raw read count and expression estimate output
- Hierarchical clustering analysis
- Clustering, PCA, heatmap correlation
- Gene body coverage
- Splicing annotation
Advanced Analysis
- All items in Standard Analysis +
- Differential gene expression
- Pathway analysis and gene ontology
Specialized Analysis
- Create a custom package to fit your project
- Custom analysis based on project requirements at an hourly rate
- TME analysis
- Fusion gene analysis
Request RNA sequencing services
Whole Genome Sequencing
High coverage, cost efficiency, and fast turnaround times
Whole genome and whole exome sequencing applications are used in a wide range of applications including the identification of clinically actionable mutations, neo-antigen prediction, and genome-wide association studies for complex diseases.
We have experience executing whole genome projects at population scale and can process diverse samples including cells, blood, tissues, and FFPE.
What library prep workflows are available for WGS?
Library
Prep
Suitable
for
Sequencing
Platform
Starting
Material
Library Prep
KAPA hyper prep
Suitable for
Any species
Sequencing
Platform
Illumina NovaSeq X+
Starting Material
DNA
50 – 1000ng
Tissue
20-40mg
Cells
>1×106
Library Prep
PacBio HiFi Prep
Suitable for
Any species
Sequencing
Platform
PacBio Revio
Starting Material
DNA
8-10ug
Tissue
Animal: 50-100mg
Plant : 1-5 grams
Cells
>3×106
What bioinformatics services are available for WGS?
Basic Analysis
- Data QC report
- Raw data FASTQ file
Standard Analysis
- All items in Basic Analysis +
- Alignment (BAM) file
- Germline VCF
- Germline variant annotation
- Low frequency germline variant calling
- Low frequency germline variant annotation
Advanced Analysis
- All items in Standard Analysis +
- Somatic mutation annotation
- Low frequency somatic variant calling
- Low frequency somatic variant annotation
- Joint genotyping
- Structural variant calling
- Tumor/normal CNV calling
- gVCF
Specialized Analysis
- Create a custom package to fit your project
- Custom analysis based on project requirements at an hourly rate
Request whole genome sequencing services
Whole Exome Sequencing
Whether you’re looking to identify medically relevant mutations, perform neo-antigen prediction, or genome wide association studies for complex diseases, we have the tailored solution to support your project from ideation to publication.
We have experience executing whole exome projects at population scale and can process diverse samples including cells, blood, tissues and FFPE.
Whole Exome Sequencing Services
What library prep workflows are available for WES?
Library
Prep
Suitable
for
Capture
Size
Input
Amount
Library Prep
Agilent SureSelect (v5, v6, v7 and v8 versions available)
Suitable for
Exome analysis of human and mouse samples from all sources including FFPE
Capture Size
48.6Mb design with 35.7Mb capture
Input Amount
200 – 500ng
Library Prep
Twist Exome
Suitable for
High-quality DNA from human and mouse
Capture Size
41.2Mb design with 36.8Mb capture
Input Amount
What bioinformatics services are available for WES?
Basic Analysis
- Data QC report
- Raw data FASTQ file
Standard Analysis
- All items in Basic Analysis +
- Alignment (BAM) file
- Germline VCF
- Germline variant annotation
- Low frequency germline variant calling
- Low frequency germline variant annotation
Advanced Analysis
- All items in Standard Analysis +
- Somatic mutation annotation
- Low frequency somatic variant calling
- Low frequency somatic variant annotation
- Joint genotyping
- Structural variant calling
- Tumor/normal CNV calling
- gVCF
Specialized Analysis
- Create a custom package to fit your project
- Custom analysis based on project requirements at an hourly rate
Request whole exome sequencing services
Single-Cell Sequencing
Generate insights at a cellular level
We are a 10X Genomics-certified single-cell sequencing service provider offering end-to-end solutions for single-cell analysis. We also developed a suite of bioinformatics tools for the analysis, interpretation and visualization of single-cell sequencing data including expanded analytical insight from heavy and light chain pairs following BCR and TCR clonotype sequencing.
We have extensive experience in single-cell gene expression analysis and multi-omics profiling using fresh cells/tissue samples, cryopreserved cells, nuclei isolated from frozen tissues and fixed cell/tissue samples. Enhanced services include cryopreservation of cells and post-submission dead cell removal to ensure the highest quality of data for your project. We also offer FACS sorting services to enrich for cell populations of interest before carrying out single cell sequencing experiments.

What workflows are available for single-cell sequencing?
Gene Expression
Explore cellular heterogeneity using 3’ or 5’ gene expression libraries for biomarker discovery and more.
Single Cell Immune Profiling
Discover full length paired sequences for T and B cells repertoire analysis.
Simultaneously measure gene expression and immune repertoires combined with cell surface protein data to look at functional states and subsets of immune cells.
Multiome Profiling
Combine gene expression with cell surface protein expression data for a multiomic view of your sample of interest.
Uncover gene regulatory programs by assaying the epigenome and gene expression simultaneously.
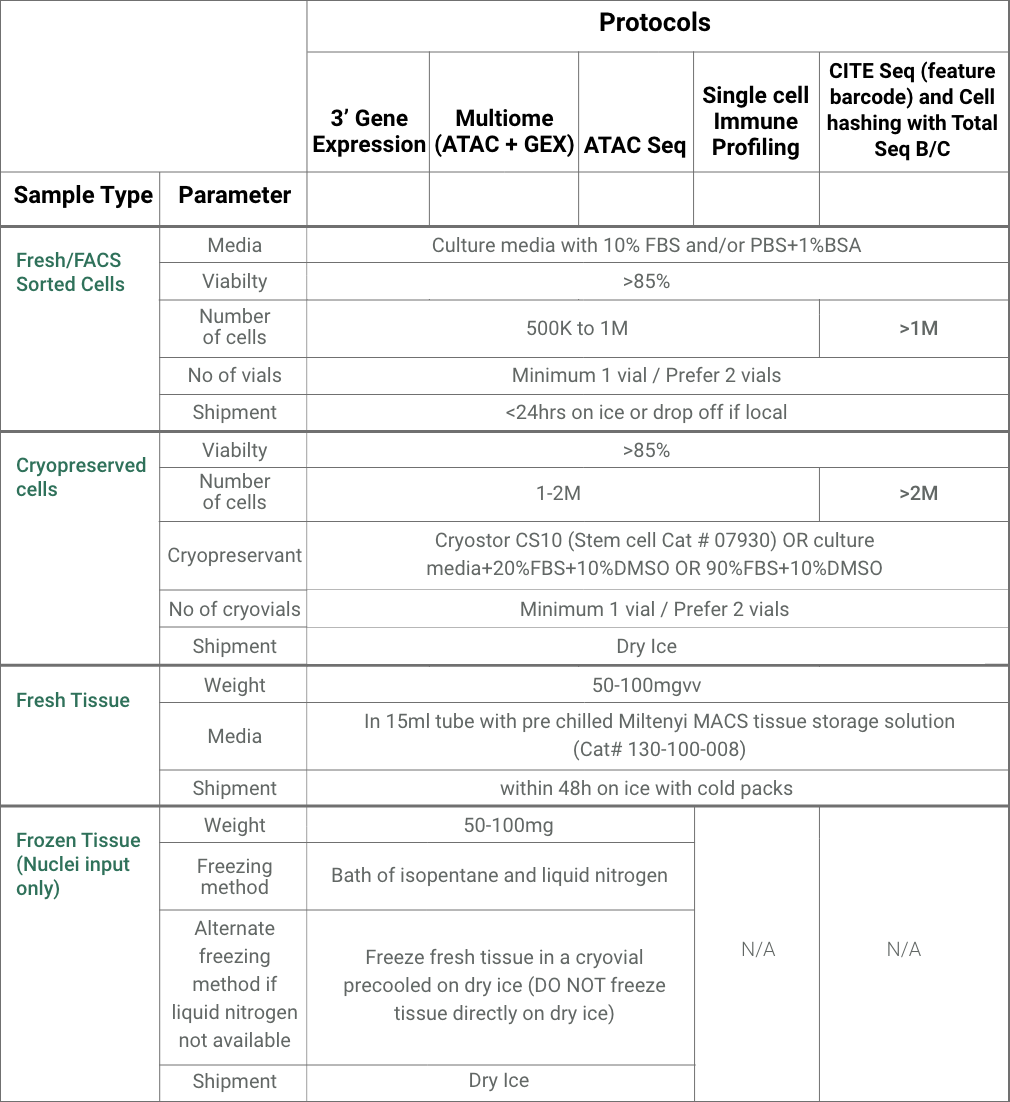
What bioinformatics services are available for single-cell sequencing?
Basic Analysis
- Data QC report
- Raw data FASTQ file
Standard Analysis
- All items in Basic Analysis +
- CellRanger outputs of sequencing QC metrics
- CellRanger outputs of gene expression and heatmap visualization
- CLOUPE file for use in LOUPE browser
- Advanced QC via Seurat
Advanced Analysis
- All items in Standard Analysis +
- Advanced filtering of low-quality cells, contamination, multiplets etc.
- Dimensionality reduction and clustering analysis on filtered data
- Interactive t-SNE plot with cluster information
- General cell type annotation
- Differential gene expression analysis for clusters and annotated cell types
- Pathway enrichment analysis
- Group comparison
Specialized Analysis
- Create a custom package to fit your project
- Custom analysis based on project requirements at an hourly rate
- Differential accessibility peak analysis for clusters and cell type in multiome projects
- Differential protein analysis for clusters and cell type in CITE-seq projects
Request single-cell sequencing services
Spatial Transcriptomics
Map gene expression profiles across tissue sections
Spatial transcriptomics technology enables high-throughput analysis of gene expression in intact tissue sections. It allows sequencing and characterization of distinctive gene expression patterns associated with different cell types that constitute a tissue while maintaining their spatial context. The resulting data can be used to identify rare cell types and gene expression heterogeneity within tissues. This is useful to study various biological processes such as development, disease pathogenesis, and tumor microenvironments.
We offer spatial transcriptome profiling of fresh frozen, fixed frozen and FFPE tissue sections.
Spatial Transcriptomics Services
What solutions are available for spatial transcriptomics?
Slide Sizes
Available
Spots Per
Capture Area
Suitable
For
Slide Sizes Available
Visium 6.5mm x 6.5mm
Spots Per Capture Area
5,000
Suitable For
Small regions of interest
Slide Sizes Available
Visium 11mm x 11mm
Spots Per Capture Area
14,000
Suitable For
Large regions of
interest
Slide Sizes Available
Visium HD 6.5mm x 6.5mm
Spots Per Capture Area
Gapless lawn of oligonucleotides
Suitable For
Single-cell scale resolution
What bioinformatics services are available for spatial transcriptomics?
Basic Analysis
- Data QC report
- Raw data FASTQ file
Standard Analysis
- All items in Basic Analysis +
- SpaceRanger generated key sequencing and gene expression metrics
- SpaceRanger generated tissue plot colored by cluster
- SpaceRanger generated t-DNA projected spot clusters
- CLOUPE file for use in LOUPE browser
Advanced Analysis
- All items in Standard Analysis +
- Advanced filtering of low-quality spots, rRNA, mitoRNA, and hemoglobin contamination
- PCA and optimized clustering of high-quality spots
- Spatial dimensional plots with filtered spots and heatmap of gene expression across clusters
- Spatial localization of individual clusters
- Cell type annotation from public databases and associated spatial plots by cell type
Specialized Analysis
- Create a custom package to fit your project
- Custom analysis based on project requirements at an hourly rate
- Cell type annotation with custom markers and associated spatial plots by cell type
- Interactome spatial visualization including number and strength of interactions
Additional services available for spatial transcriptomics projects:
- Histopathology, including H&E review and pathology assessment for quality control
- OCT Embedding
- Formalin-Fixed Paraffin Embedding
- Sectioning of FFPE blocks
Request spatial sequencing services
Immune Profiling
TCR and BCR profiling enables comprehensive mapping of the immune repertoire. It has a number of applications including understanding disease mechanisms, characterizing immunological responses, biomarker identification and development of immunotherapies. This is exemplified by the success of several immunotherapies in cancer treatment and the development of therapeutic antibodies.
Next-generation sequencing (NGS) technologies enable accurate and unbiased profiling of TCRs and BCRs with relative ease. In combination with other assays, it can serve as diagnostic or prognostic marker. We provide RNA based TCR and BCR repertoire profiling services for both bulk RNA and single cell analysis.
Immune Profiling Services
What library prep workflows are available for TCR & BCR sequencing?
Library
Prep
Suitable
for
Starting
Material
Library Prep
Takara SMART-Seq
alpha/beta with UMI
Suitable for
Full length alpha and beta chain capture for human and mouse from blood, tissue, cells and total RNA
Starting Material
Total RNA: 10 – 1000ng
Cells: 1000 – 10,000
Library Prep
Takara SMARTer BCR IgG
IgM H/K/L profiling
Full length heavy and light chain captures for human and mouse from blood, tissue, cells and total RNA
10 – 1000ng
Cells: 1000 – 10,000
Library Prep
10X 5’ Gene Expression + VDJ library TCR
Single-cell TCR analysis of blood, tissue and cells from human and mouse samples
Fresh and Cryopreserved cells
(500k – 1 million)
Library Prep
10X 5’ Gene Expression + VDJ library BCR
Single-cell BCR analysis of blood, tissue and cells from human and mouse samples
Fresh and Cryopreserved cells
(500k – 1 million)
What bioinformatics services are available for immune profiling?
Basic Analysis
- Data QC report
- Raw data FASTQ file
Advanced Analysis
- All items in Basic Analysis +
- Full-length clonotype sequences and their frequencies
- Diversity scores for each sample
- V & J gene usage
- Phylogenetic analysis of clonotypes of interest
Specialized Analysis
- Create a custom package to fit your project
- Custom analysis based on project requirements at an hourly rate
Request TCR/BCR sequencing services
Epigenomic Sequencing
DNA methylation and chromatin modifications at bulk and single-cell resolutions
What library prep workflows are available for epigenetic sequencing?
Library
Prep
Suitable
for
Input
Amount
Library Prep
ATAC-Seq
Suitable for
Chromatin accessibility studies
Input Amount
>100k
Library Prep
CUT & RUN
Suitable for
Input Amount
250k cells per antibody
Library Prep
ChIP–Seq
Suitable for
Input Amount
Immunoprecipitated DNA (>1 ng)
Library Prep
Methyl-Seq
Suitable for
Input Amount
100 – 1000ng
Library Prep
Single-Cell ATAC-Seq
Suitable for
Input Amount
> 500k – 1 million (viability >85%)
What bioinformatics services are available for epigenetic sequencing?
Basic Analysis
- Data QC report
- Raw data FASTQ file
Standard Analysis
- All items in Basic Analysis +
- Alignment (BAM) file
- Bed files
- Peak call files
- Consensus peak calling
- Hierarchical clustering analysis
- Clustering, PCA, heatmap correlation
Advanced Analysis
- All items in Standard Analysis +
- Group comparison
- Pathway analysis
Specialized Analysis
- Create a custom package to fit your project
- Custom analysis based on project requirements at an hourly rate
Request epigenomic sequencing services
Oncology Panel Sequencing
TruSight Oncology 500 offers a wide variety of benefits in analyzing multiple tumor variant types in 523 genes in a single assay. It is a next-generation sequencing (NGS) assay that enables comprehensive genomic profiling of tumor samples. The assay is highly effective in identifying all types of relevant DNA and RNA variants in different types of solid tumors including lung, melanoma, ovarian, breast, gastric, bladder, and sarcoma. Starting materials for the assay can be cells, blood, tissues, FFPE, and ctDNA with varying quality and quantity inputs.
The assay is also highly accurate in measuring immuno-oncology biomarkers such as microsatellite instability (MSI) and tumor mutational burden (TMB).
Targeted Cancer Panel Services
What library prep workflows are available for TSO 500?
TSO 500
NGS Assay
TSO 500 ctDNA
NGS Assay
TSO 500
NGS Assay
DNA & RNA
TSO 500 ctDNA
NGS Assay
Cell-free DNA
Input Amount
TSO 500
NGS Assay
50 ng purified DNA and RNA each
TSO 500 ctDNA
NGS Assay
20-30 ng purified ctDNA
Sequencing Depth
TSO 500
NGS Assay
DNA: >80M PE100 reads
RNA: >40M PE 100 reads
TSO 500 ctDNA
NGS Assay
>400M PE150 reads
What bioinformatics services are available for TSO 500?
TSO 500 DNA
- Data QC report
- Raw data FASTQ file
- Single nucleotide variants (SNVs)
- Insertions & deletions (InDels)
- Copy number variants (CNVs)
- Multi-Nucleotide variants (MNVs)
- Somatic variants
- Structural variants
- Tumor mutational burden (TMB)
- Microsatellite instability (MSI)
TSO 500 DNA & RNA
- All items in TSO 500 DNA +
- Gene fusions
- Transcript variants
- Novel transcripts
- Loss of heterozygosity (LOH)
TSO 500 ctDNA
- Data QC report
- Raw data FASTQ file
- Single nucleotide variants (SNVs)
- Insertions & deletions (InDels)
- Copy number variants (CNVs)
Request oncology panel sequencing services
Long-Read Sequencing
Unparalleled accuracy and comprehensive insights with long-read solutions on the PacBio Revio
While short-read sequencing has been an advantage for genomics study, it struggles to resolve repetitive regions, assemble complex structural variants, and fully capture isoform diversity. PacBio HiFi sequencing solves these issues with highly accurate long reads, enabling comprehensive profiling of genetic variation, including SNVs, Indels, SVs, and complex regions.

We provide both short-read and PacBio long-read solutions, supporting highly contiguous genome assemblies, structural variant detection, full-length transcript sequencing, epigenetic profiling, and haplotype phasing, providing comprehensive genomic insights for diverse research needs.
PacBio HiFi Long-Read Sequencing Solutions
What workflows are available for long-read sequencing?
Suitable
for
Sequencing
Depth
Input
Amount
WGS for Genome Assembly
Suitable for
Creating reference-quality genomes for new species
Sequencing Depth
15x per haplotype
Input Amount
3Gb genome per SMRT Cell
WGS for Variant Detection
Suitable for
Detecting large structural variants and copy number variants
Sequencing Depth
Input Amount
Kinnex Full-length Bulk RNA
New genome annotation, isoform discovery, differential isoform expression
Sequencing Depth
Input Amount
Full-length Single-Cell RNA
Suitable for
Cell-type specific, allele-specific isoform and
variant characterization in single-cell
Sequencing Depth
Input Amount
Suitable for
Targeting fragments larger than short reads can accommodate
Sequencing Depth
5-10 million reads
Input Amount
1-96 depending on coverage needed
Pure Target Sequencing
Suitable for
Targeting repeat expansion loci relevant to neurodegenerative disease
Sequencing Depth
48 plex ~800,000
24 plex ~500,000
8 plex ~225,000
Input Amount
1- 48 Multiplexed samples
What bioinformatics services are available for long-read sequencing?
Variant Detection
- Run folder with HiFi reads as aligned (BAM) file
- VCF
de novo Assembly
- Run folder with HiFi reads as aligned (BAM) file
- Report of assembly metrics
- Transgene identification (if applicable)
- FASTA file of primary contigs
- FASTA file of haplotigs
- Genome annotation (add on)
Isoform Discovery
- Report containing summary metrics for primers, reads, transcripts, transcript classifications, and more
- Full-length non-concatemer (FLNC) reads as BAM file
- FASTQ files of both low and high-quality isoforms
- Mapped high-quality isoforms as BAM file
- Collapsed filtered isoforms as GFF
Metagenomics
Full-length 16S:Targeted Sequencing
Pure-target, AAV, amplicon:Methylation (5mC) Profiling
- Run folder with HiFi reads as aligned (BAM) file
- 5mC CpG report
- HiFi reads with 5mC calls in BAM format
Request long-read sequencing services

